In order to structuralize, store and display the liver-related PPIs and their annotations which were curated from biological literature, we proposed a database named Protein-Protein Interaction Information Database (dbPPII). The structure of dbPPII is shown in Figure 1, in which a PPI Ontology (PPIO) tree view is used as a global view for the whole information of PPI data in the database.
The dbPPII is of the following benefits: First, an overview of the PPI information is given by pre-categorizing PPIs based on PPIO, a hierarchically structured vocabulary. Second, PPIs are organized into groups according to functional categories, and thus allows users to quickly navigate through the PPIs in different hierarchy levels. Third, the PPI functional sub-network which composed of PPI group is easily constructed according to the intension of user.
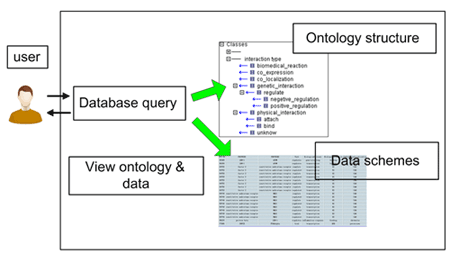
Figure 1: The scheme of the PPI annotation information
The platform was implemented using the JSP/servlet web technology. More efforts are needed to enrich the function of this database and make it more easily applicable for information capturing in the future. Currently, Human and Mouse data are available!!
Search >> |
